Heterogeneity Classifier#
This tutorial demonstrates how we can use scale-independent porosity variance to classify a given sample as homogeneous or heterogeneous.
Import packages#
from dpm_tools.io import read_image
from dpm_tools.metrics import heterogeneity_curve
from dpm_tools.visualization import plot_heterogeneity_curve
from pathlib import Path
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
/opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/site-packages/porespy/filters/_lt_methods.py:21: FutureWarning: `square` is deprecated since version 0.25 and will be removed in version 0.27. Use `skimage.morphology.footprint_rectangle` instead.
strel = {2: {'min': disk(1), 'max': square(3)}, 3: {'min': ball(1), 'max': cube(3)}}
/opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/site-packages/porespy/filters/_lt_methods.py:21: FutureWarning: `cube` is deprecated since version 0.25 and will be removed in version 0.27. Use `skimage.morphology.footprint_rectangle` instead.
strel = {2: {'min': disk(1), 'max': square(3)}, 3: {'min': ball(1), 'max': cube(3)}}
/opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/site-packages/porespy/metrics/_funcs.py:65: FutureWarning: `square` is deprecated since version 0.25 and will be removed in version 0.27. Use `skimage.morphology.footprint_rectangle` instead.
strel = {2: {"min": disk(1), "max": square(3)}, 3: {"min": ball(1), "max": cube(3)}}
/opt/hostedtoolcache/Python/3.10.19/x64/lib/python3.10/site-packages/porespy/metrics/_funcs.py:65: FutureWarning: `cube` is deprecated since version 0.25 and will be removed in version 0.27. Use `skimage.morphology.footprint_rectangle` instead.
strel = {2: {"min": disk(1), "max": square(3)}, 3: {"min": ball(1), "max": cube(3)}}
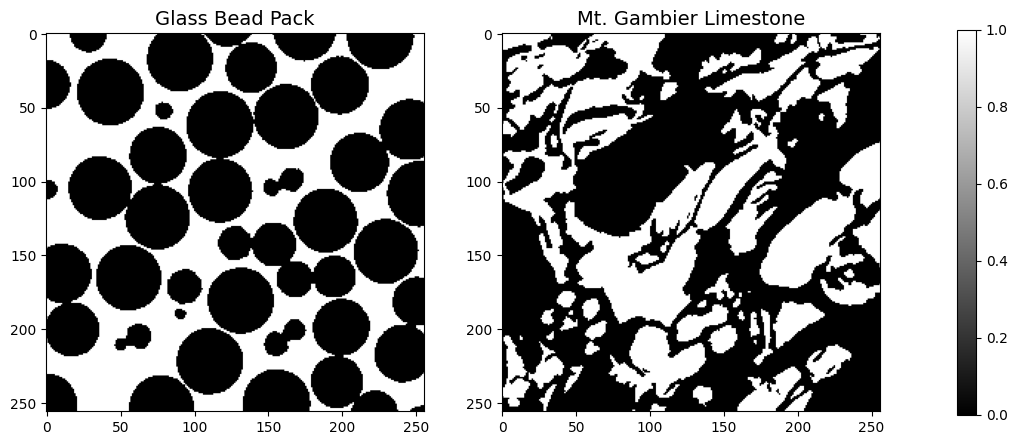
Load and visualize data#
datapath = Path('../../../_static')
beadpack = read_image(datapath / 'beadpack.tif')
gambier = read_image(datapath / 'mtgambier.tif')
fig = plt.figure(figsize=(12, 5))
gs = GridSpec(1, 3, width_ratios=[1, 1, 0.05], wspace=0.3)
# Create subplots
ax1 = fig.add_subplot(gs[0, 0])
ax2 = fig.add_subplot(gs[0, 1])
cax = fig.add_subplot(gs[0, 2])
im1 = ax1.imshow(beadpack[0,:,:], cmap='gray')
im2 = ax2.imshow(gambier[0,:,:], cmap='gray')
fig.colorbar(im1, cax=cax)
ax1.set_title('Glass Bead Pack',fontsize=14)
ax2.set_title('Mt. Gambier Limestone',fontsize=14)
plt.show()
Scale-independent Variance#
results = {'beadpack': {}, 'gambier': {}}
results['beadpack']['radii'], results['beadpack']['variance'] = heterogeneity_curve(beadpack)
results['gambier']['radii'], results['gambier']['variance'] = heterogeneity_curve(gambier)
Plot the Results#
fig, ax = plot_heterogeneity_curve(results['beadpack']['radii'], results['beadpack']['variance'], color='blue', label='Glass Beadpack')
fig, ax = plot_heterogeneity_curve(results['gambier']['radii'], results['gambier']['variance'], fig=fig, ax=ax, color='black', label='Mt. Gambier Limestone')
plt.legend()
plt.show()
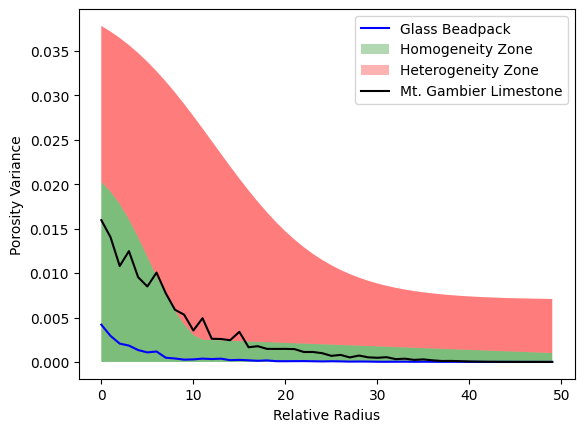
Scoring the Result#
def sigmoid(x):
"""
Sigmoid function
"""
return 1 / (1 + np.exp(-x))
def heterogeneity_score(variances) -> float:
"""
Assign a heterogeneity score to the heterogeneity curve based on the number of points above the homogeneous/heterogeneous threshold.
Parameters:
variances (np.ndarray): The porosity variance values from the heterogeneity curve
Returns:
float: The heterogeneity score for the given heterogeneity curve. 0 = homogeneous, 1 = heterogeneous
"""
x = np.linspace(-2, 17, len(variances))
bound = (0.023 * (1 - sigmoid(x)))
bnd = bound[bound <= 0.0025]
bound[bound <= 0.0025] = np.linspace(0.0025, 0.001, len(bnd))
x2=np.linspace(-2, 6, len(variances))
vv=((0.035 * (1 - sigmoid(x2)))) + 0.007
wts = vv / vv.sum()
r = wts * (variances > bound)
return r.sum()
beadpack_score = heterogeneity_score(results['beadpack']['variance'])
gambier_score = heterogeneity_score(results['gambier']['variance'])
print(f"Heterogeneity Score for Glass Bead Pack: {beadpack_score :.3f}")
print(f"Heterogeneity Score for Mt. Gambier Limestone: {gambier_score:.3f}")
Heterogeneity Score for Glass Bead Pack: 0.000
Heterogeneity Score for Mt. Gambier Limestone: 0.327